I’ve been asked several times about the best ways to include IPA characters and other “special” characters on vowel plots. Unfortunately, due to difficulties in maintaining and determining character encoding when files are transported over the web, I don’t have a good solution for this at present for the NORM website. (I’ve got some ideas here, just never seem to have the time to try them out thoroughly or to read up more on this; incidentally, this is the same reason that some of my conversion tools for Praat files, like the TextGrid to text converter, sometimes fail…)
I do have some suggestions for plotting IPA characters in Vowels though, and more generally in R, and, in fact, this can be somewhat straightforward:
R’s default behavior (at least on my Mac system and the other R installs I’ve used) is to use a font for plotting that does not support UTF-16 characters (which includes most of the IPA special characters). However, just like for other graphic parameters, you can change R’s plotting behavior to use a specified font family. Many fonts do support IPA characters. So, the simple command
par(family='Helvetica')
will tell R to use Helvetica for plot text and this will support a wide-range of characters. For example, Valerie Fridland and I did this to generate figures like the one below, figure 3, in our 2012 JPhon paper.

Without specifying the font family as Helvetica, we get this:
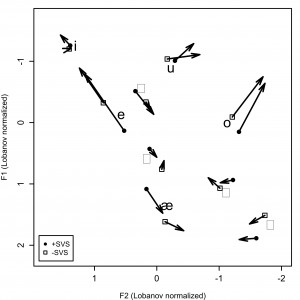
So, specifying the font family solves half the problem easily enough. However, it can also be tricky sometimes to get R to read in non-ASCII files in the first place. You can overcome this by specifying the file encoding in the read.delim() or scan() command. See
?read.delim()
And follow the help info about “fileEncoding” and “encoding”… Honesty, I find that this can still be tricky to get right sometimes, esp. when I’m reading in lots of files (with different possible encodings) through a batch processing script. I often find it easier to just store simple ASCII in my vowel data files and then to recode the vowel labels in my R script. So, continuing with the example above, to get the IPA labels in the first place, I just do something like this… (Before this point, I’ve loaded all of my vowel data (for three regional groups, “Northerners”, “Southerners”, “Westerners”), normalized it (here via Lobanov), and then extracted just the “Southern” data and stored it in the data frame called “svowels”.)
> levels(svowels[,2]) # the vowel labels are just stored in a BvowelT format
[1] "BAIT" "BAT" "BEAT" "BET" "BIT" "BOAT" "BOOT" "BOT" "BUT"
> levels(svowels[,2]) <- c("e", "æ", "i", "ɛ", "ɪ", "o", "u", "ɑ", "ʌ")
> levels(svowels[,2]) # now the labels are in IPA, note must be careful \n
about the order, matching the order in the original levels
[1] "e" "æ" "i" "ɛ" "ɪ" "o" "u" "ɑ" "ʌ"
> jpeg(filename = "~/Documents/Samples/JPhon_fig3_example.jpg", width = 90, \n
height = 90, units = "mm", res = 1000, pointsize = 6, \n
quality=100, bg = "white", type = "quartz")
> par(mai=c(0.4, 0.4, 0.1, 0.1), family='Helvetica')
> vowelplot(compute.means(svowels, separate=T), \n
color=NA, label="none", title=" ", l.size=1)
> text(compute.means(svowels, separate=F)[,5], \n
compute.means(svowels, separate=F)[,4], \n
as.character(compute.means(svowels, separate=F)[,2]), \n
pos=c(2, 3, 4, 4, 1, 1, 4, 2, 4), cex=2)
> dev.off()
null device
1
A few words about this. The lines with the > are lines executed in R. I’ve indicated line-wraps with “\n”, the new line character (I’ve wrapped these since otherwise they run off the side of the screen). Lines that begin with [1] and the two lines at the bottom “null device” and “1” are output from R.
Since the second column of a vowel data.frame, where the vowel label is stored, is interpreted by R as a factor, the levels() command will both output these levels and allow you to replace them. So, the first instance,
> levels(svowels[,2]) # the vowel labels are just stored in a BvowelT format
[1] "BAIT" "BAT" "BEAT" "BET" "BIT" "BOAT" "BOOT" "BOT" "BUT"
shows us what values exist in this column. The second instance,
> levels(svowels[,2]) <- c("e", "æ", "i", "ɛ", "ɪ", "o", "u", "ɑ", "ʌ")
replaces those levels with IPA versions. It’s very important to have the order right! Then the third,
> levels(svowels[,2]) # now the labels are in IPA, note must be careful \n
about the order, matching the order in the original levels
[1] "e" "æ" "i" "ɛ" "ɪ" "o" "u" "ɑ" "ʌ"
let’s us see that we have set them correctly. (Compare this output to the output above from the first levels() command.)
I won’t dwell on this here, but the jpeg() command tells R to send the graphical output to a jpeg file named “~/Documents/Samples/JPhon_fig3_example.jpg” and allows us to specify a lot of detail. The par() command lets us specify the font family, the crucial step for plotting the IPA characters, but we also use it to adjust the margins of the figure.
I’m using the normal vowelplot() command to generate the basic plot, but then note that I’m using R’s standard text plotting function, text(), to add the actual labels. This isn’t strictly necessary – once we’ve specified a UTF-16 supporting font family, the vowelplot() function will get the labels right – but I’ve included this here as a demonstration that we can use the vowelplot() function to set up the plotting space and then we can use other R functions to annotate that space. The text() function gives us a lot more control over where the labels go so I’ve used it here. See
> ?text
to see what I’m doing with the “pos” parameter, if you’re not familiar with it. Finally, dev.off() tells R to finish writing to the file.
There’s a lot more we can do to customize vowel plots in R, but I’ll save other sample code and suggestions for later posts. Please feel free to leave a comment or send an email if any of this isn’t clear, or if you’d like me to comment more on anything. I hope this is helpful!


