The overarching goal of the Harms lab is to understand the relationship between the biophysical properties of proteins and their evolution. Why do proteins with certain sequences and physical properties—out of a huge space of possibilities—occur? How do the physical properties of proteins shape their evolutionary trajectories? Which protein features are optimized by evolution, and which are determined by chance? How does a blind evolutionary process assemble complex features like ligand binding sites or allosteric regulation? Is protein evolution predictable or stochastic? To answer these (and other) questions, we take a synthetic approach, combining concepts and methodologies from classical biophysics and evolutionary biology. We employ advanced phylogenetics techniques (including ancestral protein resurrection), high-throughput experimental screens, and rigorous experimental/computational biophysical approaches to directly study the interplay of evolutionary and biophysical forces in generating both the complexity and diversity of natural proteins.
Exploring sequence space
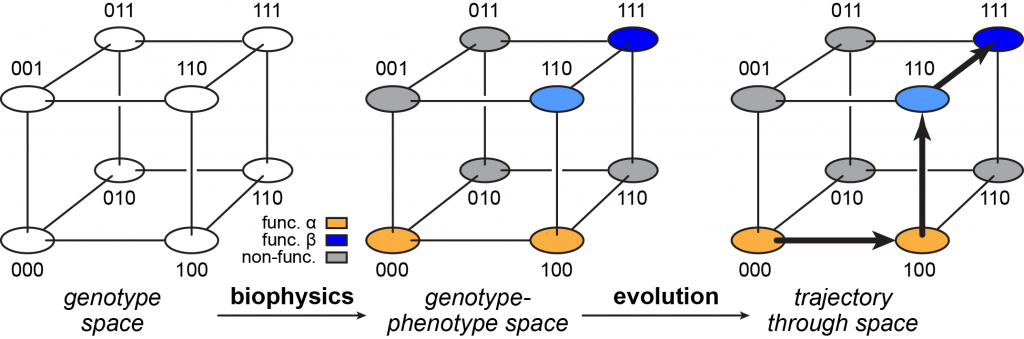
Sequence space provides a rich metaphor to organize thinking about the evolution of proteins and reveal the common ground shared by evolutionary biology and biophysics. The simplest sequence space is a “genotype space” that contains all possible amino acid sequences and the mutational connections between them. Each sequence is a node, and each node is connected by edges to all neighboring proteins that differ from it by just one amino acid. The genotype space becomes a “genotype-phenotype” space when each node is assigned information about its functions. Finally, as they evolve, proteins follow trajectories along edges through the genotype-phenotype space.
Biophysics and evolutionary biology have traditionally addressed different aspects of this map. Biophysicists have sought to characterize the map’s structure and its physical determinants —the links among protein sequence, biophysical properties, and function. Evolutionary biologists have studied the trajectories that proteins follow through this map and the evolutionary forces that drive them to do so. We unite these approaches, seeking to reveal how and why proteins evolve across genotype-phenotype space to produce the diversity of proteins found in nature.
Figure 1. Sequence space. The left panel shows a simple genotype space. Each possible sequence is a node. Neighboring nodes that differ by only one point mutation are connected by edges. The example shows a three-site protein with only two possible states (0 or 1). The middle panel shows genotype-phenotype space, where each sequence is associated with its functional characteristics, which are determined by the molecule’s biochemical properties. Here, three possible states are shown: function α (orange), function β (blue), and non-functional (gray). An intermediate state between α and β is shown in light blue. Evolutionary processes drive proteins across the genotype-phenotype space. The right panel shows one trajectory beginning at genotype 000 and ending at 111
The S100 protein family as an evolutionary biophysical model
A powerful model system allows deep and nuanced studies that provide insights inaccessible in more complex systems: Drosophila for evolutionary developmental biology, ribonuclease H for protein folding, and—in our case—the S100 family for evolutionary biophysics. The S100s are small (~10 kDa) allosteric calcium binding proteins that ligate calcium and then recruit and regulate specific target proteins. They possess a number of properties that make them an excellent family for asking evolutionary biophysical questions.
- The S100s are functionally and biophysically diverse. Humans possess 21 family members that are involved in a wide variety of cellular processes including the stress response, cell motility, signaling, and tumor suppression. They are important for organ and tissue development, inflammation, and antimicrobial defense and have been implicated in autoimmune disease, cancer, and neurodegenerative disorders. This functional diversity is undergirded by biophysical diversity, including altered metal binding, protein target binding specificity, binding cooperativity, allostery, and oligomerization state. By studying how these core biophysical properties evolved in the S100s, we gain insight into how these properties evolve in other protein families.
- The S100s are experimentally and phylogenetically tractable. A key feature of a model system a match between what is asked and what can be studied experimentally. The S100s can be easily expressed/purified and are well behaved in solution, making them amenable for biophysical characterization. Further, the shared properties of the protein family mean that early experimental development for studies of a few family members lower the barrier for future studies of interesting evolutionary transitions across the protein family. They are also small enough that the entire protein sequence can be covered with Illumina paired-end reads, allowing high-throughput studies of mutations at any (or all) sites in the protein. Finally, they align well and possess enough phylogenetic signal to allow robust phylogenetic inference and high quality reconstruction of ancestral protein sequences.
- Current S100 projects. We are currently using S100 family members to ask how allosteric sites can evolve de novo. One family member acquired a new, an antagonistic binding site ~300 million years ago. How could a blind process assemble a site with multiple residues? Were there functionally neutral—or even deleterious—steps on the way? To what extent is the allostery optimized rather than a “natural” consequence of the protein architecture? We also have other projects in the pipeline looking at properties like the evolution of heterodimeric proteins from homodimeric ancestors, and the convergent evolution of peptide binding sites.
Methodology
We employ a variety of methods to explore protein sequence space and study the interplay between its properties and the historical evolutionary process.
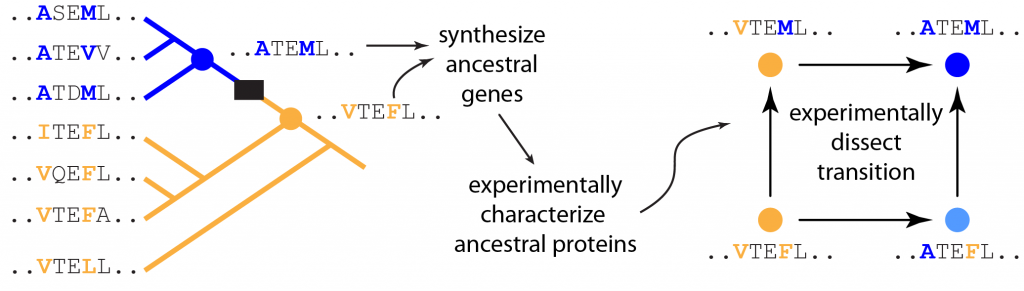
- Phylogenetics/ancestral protein resurrection. Evolution has been a massive experiment, conducted in parallel over billions of years, in the diversification and optimization of protein structure and function. The data from this experiment persist in the patterns of conservation and variation in present-day sequences; explicitly evolutionary analysis provides the means to directly interpret those data. One of the most powerful approaches in our toolkit is to “resurrect” key ancestors that bracket a change in protein function using the phylogenetic technique of Ancestral Protein Resurrection (APR). We statistically estimate the sequences of ancestral proteins, and then synthesize and experimentally characterize these ancient proteins. In contrast to “horizontal” comparisons between modern proteins, this “vertical” approach efficiently isolates a small number of sequence changes correlated with a functional transition and allows experimental characterization of the effects of historical substitutions in the relevant historical sequence background. Knowledge of what happened historically provides an important baseline for further studies of “might-have-been” alternate evolutionary trajectories.
Figure 2. Ancestral protein resurrection. Color represents ancestral (orange) or derived (blue) protein function. Black box highlights the evolutionary interval over which the transition occurred. Modern protein sequences are shown on the left, two ancestral sequences are on the right. Highlighted residues changed over the interval of interest. These genes can then be synthesized and their products experimentally characterized. Further, the effects of the historical mutations can be measured in the ancestral protein background.
- High throughput experimental characterization of sequence space. Evolutionary trajectories are strongly shaped by the distribution and connectivity of protein function across sequence space. This has been difficult to study experimentally because of the large volume of sequence space; however, recent advances in high throughput sequencing have enabled new approaches to experimentally characterize sequence space. For example, using phage display coupled to Illumina sequencing, we can simultaneously measure the binding properties of millions of clones. We are developing robust computational tools to analyze these data and explore how different evolutionary scenarios—natural selection, neutral drift, etc.—could drive evolution through this space.
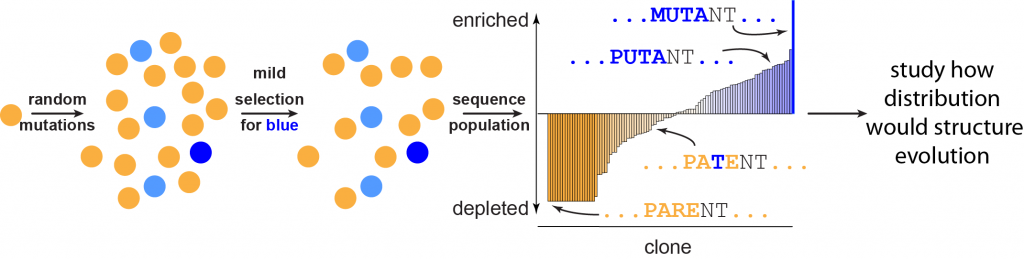
Figure 3. High-throughput explorations of sequence space. An initial protein (orange) is subjected to random mutagenesis. Weak selection for a function of interest is then applied, preferentially enriching clones possessing the function and depleting those that do not possess it. The entire population is then sequenced using high-throughput technology. By measuring the frequency of each clone in the final population, the phenotypes of millions of clones can be simultaneously characterized.
- Computational explorations of sequence space. The sheer volume of protein sequence space—a 100 amino acid protein has 20100 (~1×10130) possible sequences!—means that most of it will remain forever inaccessible to experimental studies. To gain insight into this space, we are developing novel computational approaches that leverage phylogenetic information and data from our high-throughput experiments to estimate the properties of these experimentally inaccessible volumes of sequence space.
- Rigorous biophysics. Ultimately, we want to understand the properties of sequence space in physical chemical terms. While high-throughput studies reveal correlations between genotype and phenotype, establishing causal relationships requires rigorous, manipulative biophysical investigations. We use a variety of techniques to characterize ancestral proteins, the effects of important historical mutations, and dissect key features of sequence space revealed by our high-throughput studies. We measure binding thermodynamics using isothermal titration calorimetry; measure thermodynamic stability by chemical denaturation; study solution structure and dynamics using fluorescence, circular dichroism, and electron paramagnetic resonance spectroscopy (and, soon, NMR); obtain structural information with X-ray crystallography; and use molecular dynamics simulations to interpret and extend our experimental results.
Conclusion
The why of proteins lies in the interplay of historical and physical causes, and only a mode of explanation that incorporates both types of analysis can provide a complete understanding of that interplay. By employing a wide variety of phylogenetic, experimental, and computational techniques, members of the Harms lab are working to define the physical determinants of the space over which sequences evolve and characterize the evolutionary processes that produce proteins’ diverse physical properties. Ultimately, we aim to transcend interdisciplinary barriers and treat proteins as integrated physical and historical wholes.