Ken-ichi Noma
(Details)
Professor of Biology
Member of : Institute of Molecular Biology
Ph.D., University of Tokyo
B.S., Nagaoka University of Technology
Research Interests
- 3D Genome Organization
- Chromosome Dynamics
- Transcription
- Epigenetics
- Cancer
- Aging
- Cellular Senescence
- Yeast and Human systems
Overview
Eukaryotic genomes are non-randomly organized in the nucleus. Instead, they are organized into a three-dimensional genome structure that participates in various nuclear processes such as transcriptional regulation, DNA replication and repair, as well as chromatin domain formation. Disorganization of this structure is correlated with human disease, including cancer. Despite the clear importance of 3D genome organization to basic and medical research fields, genome-organizing mechanisms and their functions in distinct nuclear processes are poorly understood.
The Noma laboratory has studied 3D chromosome organizations in the fission yeast model organism and the more complex human system. To pursue these studies, the Noma laboratory employs genomic technologies and single locus/live-cell imaging technology along with molecular and chromatin biology and epigenetics.
The Noma laboratory seeks to determine evolutionarily conserved 3D genome-organizing mechanisms and their functions in the yeast model as well as in the human system, and also establish a clear mechanistic framework for understanding the molecular underpinnings for human cancers, aging, and cellular senescence, all of which involve the reorganization of the cell nucleus.
Deciphering 3D genome structure
We have modeled the 3D genome structure of the model organism fission yeast using a genomic approach that combines the molecular biology procedure called chromosome conformation capture (3C) and next-generation DNA sequencing. To accurately model this genome structure, we successfully fused microscopy and genomic data (Tanizawa et al. Nucleic Acids Research 2010).
Bridging Transcription and 3D Genome
We have identified a molecular mechanism whereby the dividing cell protects and faithfully segregates actively transcribed genes. Specifically, we have shown that the condensin subunit Cnd2 binds directly to the most important general transcription factor, the TATA box-binding protein TBP, and that TBP recruits condensin molecules onto RNA polymerase III-transcribed genes and highly active Pol II-transcribed genes (many ribosomal protein genes) through the Cnd2-TBP interaction (Figure A). Condensin in turn tethers these highly active genes to centromeres, such that they are protected and faithfully segregated during mitosis. We found that the interaction between two classic factors, condensin and TBP, plays a pivotal role in 3D chromosome organization, and ensures that a critical set of genes, the actively transcribed housekeeping genes, are accurately segregated during mitosis (Figure B and C). This mechanism coordinates transcription with chromosomal architecture by linking dispersed gene loci with centromeres. This study was published in Molecular Cell (Iwasaki et al. 2015).

(A) The interaction between the condensin complex and TBP mediates clustering of Pol III-transcribed genes (tRNA) and highly active Pol II-transcribed genes at centromeres. (B) In wild-type cells, those highly active genes are tethered to centromeres, and chromatin loops derived from centromeres are supercoiled. This chromosomal arrangement efficiently transmits mechanical force pulling the kinetochore to chromosomal arm regions, thereby improving the fidelity of chromosomal segregation. (C) In condensin mutant, highly active genes do not associate with centromeres. Therefore, physical force at the kinetochore is diffused and inefficiently transmitted to chromosomal arm regions, resulting in chromosomal segregation defects in condensin mutant.]
Actions of Condensin and Cohesin in 3D Genome Organizations
How condensin and cohesin govern the 3D structure of the eukaryotic genome is an exciting research area. Using an important genomic methodology, referred to as ChIA-PET (Chromatin Interaction Analysis by Paired-End Tag sequencing), we have been able to capture condensin- and cohesin-mediated gene contacts throughout the fission yeast genome. Our results have revealed that although condensin and cohesin bind to the same gene loci, they direct different association networks (Figure). Cohesin mediates local contacts, i.e. between loci positioned within 100 kb (red), whereas condensin drives longer-range contacts (blue). Cohesin forms small topological chromatin domains of approximately 100 kb, while condensin organizes 300 kb – 1 Mb domains. These studies demonstrate that the two important protein complexes, condensin and cohesin, are both essential for the assembly of the functional genome architecture, but their roles in the 3D genome organization (gene contacts and topological domain organization) are significantly different. This study has been published in Nature Genetics (Kim et al. 2016).
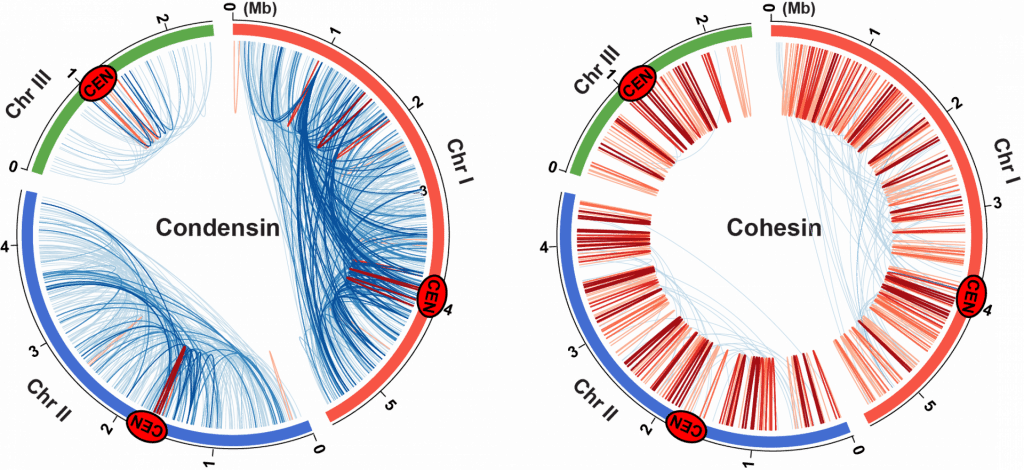
ChIA-PET genomic analyses successfully mapped condensin (left) and cohesin (right)-mediated gene contacts throughout the fission yeast genome. Blue and red lines represent gene contacts between two gene loci positioned more than and less than 100 kb away, respectively.
Visualizing associations between discrete genomic regions in live cells
We have employed a live-cell imaging system to visualize the genomic region carrying lacO repeats. We have integrated lacO repeats into the genomic region of interest and introduced LacI-GFP proteins by genetic crosses. We have been able to visualize several diverse genomic regions: centromeres, telomeres, ribosomal DNA (rDNA) repeat locus, and loci carrying retrotransposons or Pol III-transcribed genes including tRNA and 5S rRNA, in live fission yeast cells. Using a laser scanning confocal microscope, we have examined positions of the genomic loci in the 3D nuclear compartment at as short as 1.5-second intervals. Importantly, these studies to elucidate gene contacts in the 3D nuclei of live cells have demonstrated that condensin-mediated contacts between centromeres and the genomic loci carrying Pol III-transcribed genes or retrotransposons are highly dynamic. Furthermore, our results have revealed that centromeric motion, which is controlled by cytoplasmic microtubules, contributes to the mobility of non-centromeric gene loci. We propose that centromeres serve as “Genome Flexibility Elements” by connecting dispersed gene loci to highly mobile centromeres (Kim et al. Journal of Cell Science 2013).
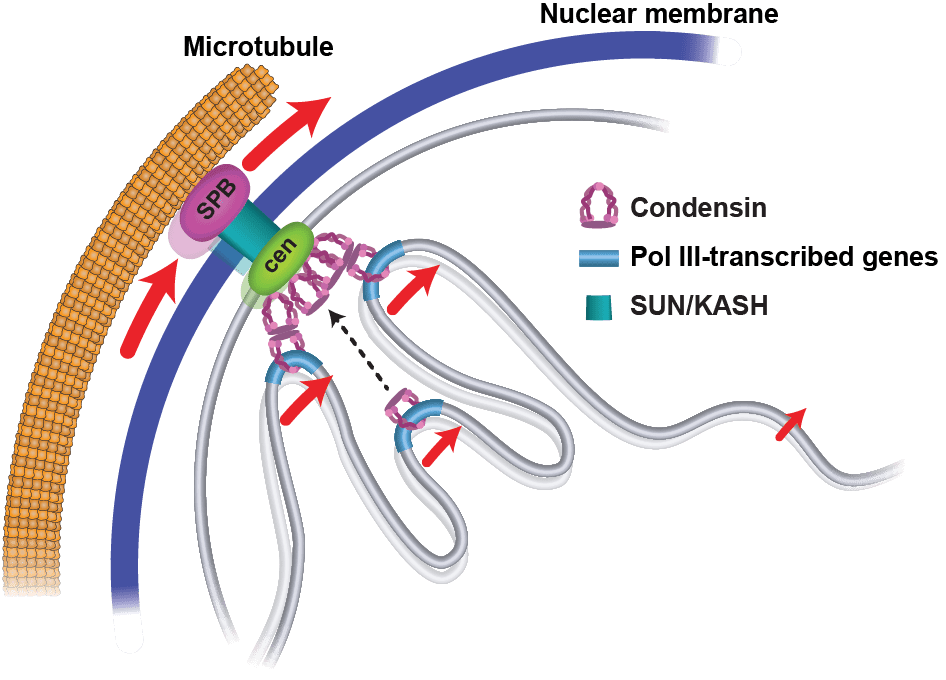
A model for the role of centromeric motion in the mobility of many gene loci during interphase. Microtubule polymerization in cytoplasm actively pushes centromeres in nucleoplasm. Centromeres associate with dispersed gene loci, and these gene contacts are mediated by condensin. Centromeres and their associating chromosomal loci migrate in a coordinated fashion.
3D Genomic Basis of Cellular Senescence
Cellular senescence is defined as a state of stable cell growth arrest, and it is our intrinsic defense against abnormal cell proliferation associated with every type of cancer. The process of senescence requires global alterations in the genome architecture and radical changes in gene expression. We have shown that the human condensin complex functions in global 3D genome reorganization during the important process of cellular senescence (Yokoyama et al. Cell Cycle 2015).

Condensin triggers cellular senescence and its associated genome organization. Condensin expression in non-senescent human cell (left) induces senescence and completely transforms global genome architecture (right), which is referred to as Senescence-Associated Heterochromatic Foci (SAHF). DNA was visualized by DAPI.