Single Molecule Biophysics
Quantitative analysis of membrane signaling event with single molecule resolution
Biochemical characterization of most membrane signaling reactions has been studied almost exclusively using bulk solution assays with either lipid head groups or lipid vesicles as substrates. These solution-based measurements have provided a foundational understanding of some membrane signaling reaction. However, this general approach suffers from poor temporal resolution and lacks information about the spatial organization and dynamics of membrane synthesis reactions. Being able to quantitatively measure the relationship between the membrane density of signaling inputs and the corresponding enzyme activity is critical for understanding how these enzymes are regulated in different cell types and signaling pathways. To better understand the relationship between lipid modifying enzyme activity and the membrane density of stimulatory molecules, we have developed fluorescence imaging methods to visualize how membrane signaling reactions are activated and inactivated with single molecule resolution using Total Internal Reflection Fluorescence (TIRF) Microscopy. We can now quantify single molecule dwell times, diffusion coefficients, and enzyme catalysis on supported lipid bilayers using purified proteins.
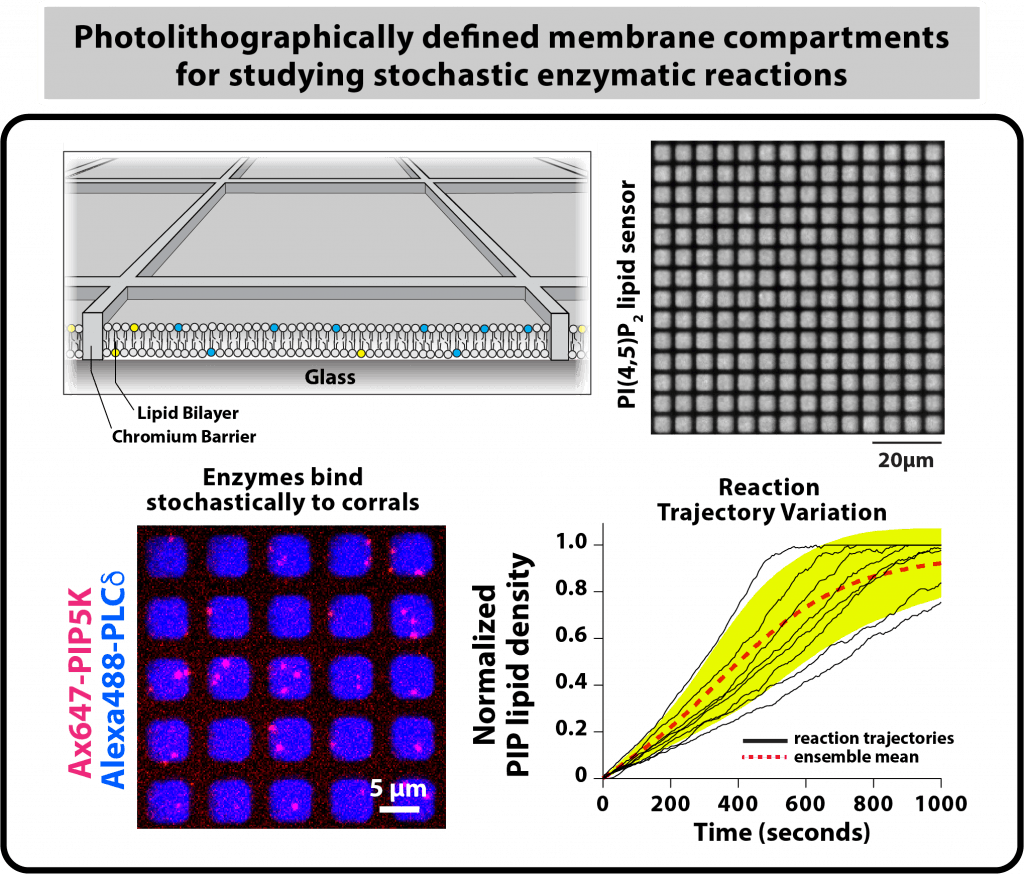
Micropatterned membranes for measuring single enzyme kinetics
In cells, proteins function in geometrically complex environments where molecular copy numbers stochastically fluctuate and the surface-area-to-volume ratio of cellular membranes constantly changing. My lab aims to biochemically reconstitute signaling reaction that mimic the organization found in living cells, rather than a test tube. We are also interested in understanding how reaction outcomes are influences by stochastic membrane binding of signaling molecules that are present at low molecular copy number in cells. Working in the Center for Advanced Material Characterization in Oregon (CAMCOR), my group has established methods to fabricate substrates that can be used to biochemically reconstitute membrane signaling reactions using physiologically relevant boundary conditions that mimic spatial confinement seen in cells. Specifically, we established a photolithography method for micropatterning supported lipid bilayers with chromium barriers. This method allows us to partition membranes into hundreds of individual membrane reaction compartments, which spatially isolates PIP lipid substrates and products from neighboring membranes. This experimental approach allows us to directly visualize the catalytic output of single lipid modifying enzymes tethered to membrane surfaces.